How to maximize the power of your 10X Genomics single-cell and spatial data
Get the most out of your data with Platforma
Introduction
10x Genomics’ Universal 5’ Gene Expression products generate full-length, paired V(D)J sequences from individual cells, providing rich insights into the clonal diversity of immune receptors. While Cell Ranger and Loupe Browser are powerful tools for analyzing 10x Genomics data, incorporating MiXCR into your workflow can unlock even deeper immunological insights.
MiXCR comes equipped with advanced computational capabilities, allowing you to:
Perform advanced error correction for PCR and sequencing artifacts
Detect cross-cell contamination
Support individual or strain-specific allele inference
Enable construction of B cell somatic hypermutation trees
Integrate with bulk RNA-seq and repertoire sequencing data
Support a broad range of species beyond human and mouse
Accommodate custom transcriptome references
Facilitate analysis of unconventional immune chains, including gamma delta (γδ) TCR repertoires
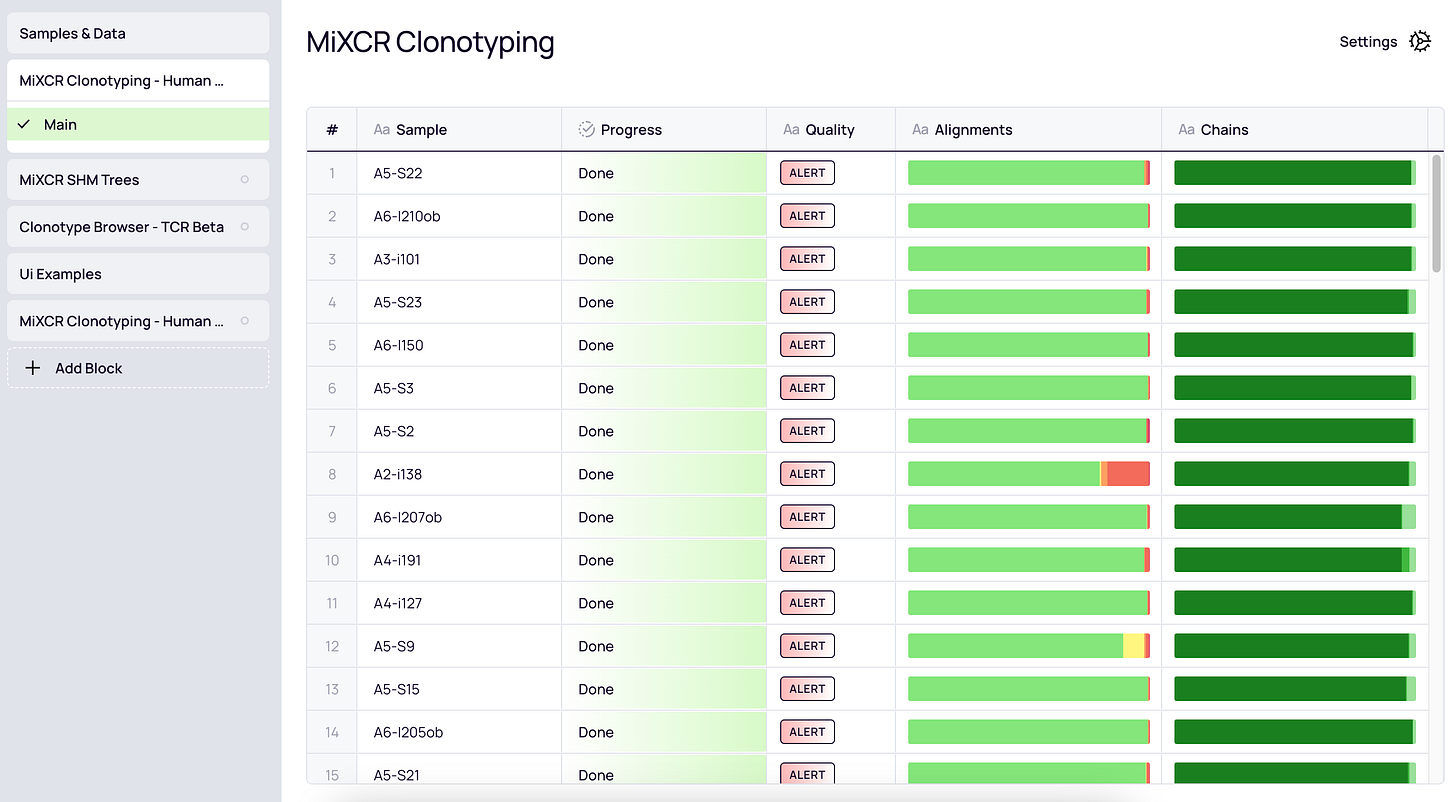
MiXCR is available as a command-line tool, or as a block on Platforma - UI‑first bioinformatics platform designed for analysis and visualization of sequencing data without any coding (figure below). Either option allows you to analyze 10x Genomics Universal 5’ V(D)J data with simple, ready-to-use presets on a laptop, server, or in the cloud.
MiXCR workflow
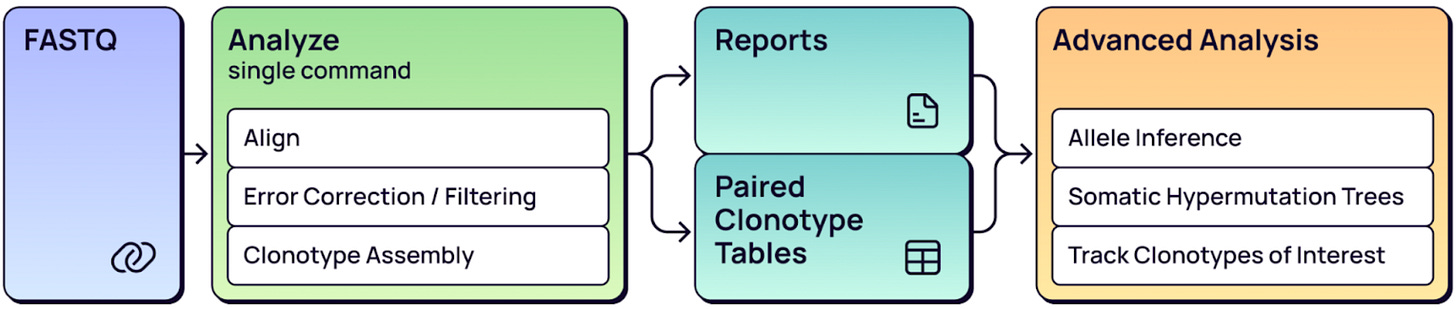
The MiXCR workflow consists of three parts:
Part 1: Upstream analysis
Assemble contigs - Longer consensus chains are assembled from short reads in the FASTQ files.
Alignment - Consensus sequences are aligned to the reference.
Filtering - Alignments are filtered based on reads per UMI and UMI per cell distributions, lowering artificial diversity.
Sequencing and PCR error correction - Multiple error correction steps reduce PCR and sequencing errors.
Clonotype assembly - Full-length clonotype sequences are reconstructed.
Chain pairing refinement - Cross cell contamination is removed and multiplets are resolved.
Part 2: Quality control
QC report generation - Comprehensive text reports are created for every step of the pipeline.
QC plot generation - Percent alignment, chain usage, and UMI/cell barcode distribution plots assessing quality of one or more samples are created.
Part 3: Downstream secondary analysis
Somatic hypermutation trees generation
Allele inference
Link metadata to samples
CDR3 characteristics analysis: strength, hydrophobicity, volume, charge, and more.
Multiple diversity measures: Normalized Shannon-Wiener, Chao1, Gini Index, and more.
Segment usage analysis
Pairwise distance analysis
The MiXCR workflow starts with the raw sequencing data, usually a FASTQ file from your sequencing vendor of choice. MiXCR is equipped to analyze all 10x Genomics VDJ chemistries, including the GEM-X (5’ v3) chemistry - just select the already available 10x Genomics Preset. Upon initiation, MiXCR performs the upstream analysis (Part 1) and QC steps (Part 2) as summarized above. It then generates QC reports and a list of clones for paired TCRα/β or IG heavy/light chains. Afterwards, you can proceed with the downstream secondary analysis (part 3).
Advantages of MiXCR
MiXCR is the leading software for immune repertoire analysis, trusted by 47 of the top 50 research institutions and 8 of the top 10 biopharma companies, thanks to its ease of use, comprehensive features, exceptional speed, and high accuracy. It has been used to analyze over 10 million samples and is cited in more than 1,700 academic publications.
Easy to use
Performing secondary analysis on 10x Genomics Universal 5’V(D)J data often requires the use of multiple tools and coding proficiency. However, using MiXCR on Platforma streamlines the entire process, including secondary analyses, into drag-and-drop blocks. Use dedicated presets for 10x Genomics: mixcr analyze 10x-sc-xcr-vdj or mixcr analyze 10x-sc-xcr-vdj-v3, making analysis simple and accessible even to those without coding skills.
Rich with features
MiXCR offers many analysis features to process all on market bulk or 10x Genomics Universal 5’ V(D)J data. Beyond core functionalities, here are some features you can use in MiXCR:
Multimodal analysis: Integrate bulk, single cell, or Sanger data.
Custom reference library: Align data to IMGT database, built-in curated library, or custom-made library.
Supports many species: Analyze data from nonstandard species using custom library.
Secondary analysis options: Take advantage of a comprehensive set of secondary analysis tools.
Fast processing speeds
Computational efficiency is crucial for immune system profiling, particularly when handling large single cell datasets. In benchmarking tests, MiXCR was faster, more sensitive, and more accurate than other popular tools like TRUST4 and Immcantation. Additionally, MiXCR allows for easy scalability of resource usage (e.g., CPU and memory) as per your data processing needs.
Highly accurate
Accuracy is paramount when working with complex immune sequencing data. MiXCR delivers highly accurate results with negligible error rates, even in the most challenging samples. The MiXCR analysis pipeline incorporates multiple error correction steps that eliminate errors from PCR, sequencing, and UMI artifacts. You can see examples of MiXCR’s accuracy in the benchmarking tests.
Getting started with MiXCR
MiXCR is available as a block (application) on Platforma. To get started, download Platforma (free for academia). You can also visit the Platforma Getting Started page, which walks you through multiple deployment options.
Requirements
Compatible with all Mac, Linux, and Windows. The minimum configuration to run on the desktop is 8 GB of RAM, 4 CPU cores, and 20 GB disk space.
Support
To ask questions and interact with the Platforma community, check out our community forum or speak directly to the support team at support@milaboratories.com.